What'sNEW June–July 2008
 20 July 2008 20 July 2008
The number of cases of HGT involving eukaryotes is now so great that it no longer makes sense to attempt to comprehensively compile individual cases — Patrick Keeling and Jeffrey Palmer
|
Analysed in isolation, a gene transferred between distant species [or into both!] will make the species seem closely related.
|
In a broad new survey, Keeling and Palmer discuss gene transfers to eukaryotes from organellar genomes, symbiotic prokaryotes, nonsymbiotic prokaryotes, and other eukaryotes. One transfer between fungal species apparently occurred only 70 years ago. In another instance, a species of green algae apparently acquired a whole chromosome by transfer. In yet another, the entire genome of a common intracellular bacterial endosymbiont, Wolbachia is found in the genome of a fruitfly. But Keeling and Palmer are actually quite conservative in estimating the importance of horizontal gene transfer in eukaryotes, for several reasons. They write —
- The gold standard for identifying HGT with confidence is …strong conflict between the phylogenies of the gene and of the organism. [So HGT between related species will be overlooked.]
- To confidently detect phylogenetic incongruence requires a level of taxonomic sampling for the gene in question that is often lacking at present…. [So many HGTs between unrelated species will be overlooked.]
- The more ancient a transfer, the harder it is to detect. [So ancient ones will likely be overlooked.]
- An organism with traits that seem to discourage HGT today might have evolved from ancestors with traits that encouraged it, and it might still harbour many genes acquired by that ancestor. [More ancient transfers will go unnoticed.]
- Eukaryote–eukaryote transfer of nuclear genes is underestimated for a number of reasons.... Given all this, it is noteworthy how many gene phylogenies have led to the conclusion that genes are in fact transferred between eukaryotes.
- Most described cases of HGT in eukaryotes involve bacterial genes.
- ...Looking forward from the many transfers that are already known to be a source of new functions, it is clear that HGT should feature prominently in future studies on the comparative genomics of eukaryotes.
- The ecological advantages that HGT might confer during a period of adaptation, such as becoming anaerobic or parasitic, are obvious (and this is not restricted to bacterial genes), but this is not to imply that HGT is only associated with extreme adaptations.
The importance of HGT in evolution is a basic prediction of cosmic ancestry. It comes as a surprise for darwinism. The surprise is especially jarring when transferred genes arrive with functions for which their donors have no obvious use. (For example, see our entry next below). Nonetheless, in a silent paradigm shift, HGT among prokaryotes has become the accepted rule. And now HGT into and among eukaryotes is being recognized. We hope the implications of this possible paradigm shift will not be ignored.
 Patrick J. Keeling and Jeffrey D. Palmer, "Horizontal gene transfer in eukaryotic evolution" [abstract], doi:10.1038/nrg2386, p 605-618 v 9, Nature Reviews Genetics, Aug (online 1 Jul) 2008. Patrick J. Keeling and Jeffrey D. Palmer, "Horizontal gene transfer in eukaryotic evolution" [abstract], doi:10.1038/nrg2386, p 605-618 v 9, Nature Reviews Genetics, Aug (online 1 Jul) 2008.
 the Keeling Lab, Canadian Institute for Advanced Research, Evolutionary Biology Program, University of British Columbia, Vancouver, BC. the Keeling Lab, Canadian Institute for Advanced Research, Evolutionary Biology Program, University of British Columbia, Vancouver, BC.
 Viruses and Other Gene Transfer Mechanisms is our main CA webpage about gene acquisition Viruses and Other Gene Transfer Mechanisms is our main CA webpage about gene acquisition
[ What'sNEW about HGT What'sNEW about HGT  ] ]
 Bats and horses are closely related — a likely error caused by ignoring HGT, What'sNEW, 20 Jun 2006. Bats and horses are closely related — a likely error caused by ignoring HGT, What'sNEW, 20 Jun 2006.
 ...Genomic complexity was present very early on....: many advanced Trichoplax genes have no obvious use in Trichoplax, What'sNEW, 22 Aug 2008. ...Genomic complexity was present very early on....: many advanced Trichoplax genes have no obvious use in Trichoplax, What'sNEW, 22 Aug 2008.
 10 July 2008 10 July 2008
 We don't have a clue — Gerard Manning of the Salk Institute, answering the question, "What is a single-celled organism doing with all this communications gear?" We were absolutely stunned. Manning's group is another one studying Monosiga brevicollis, found to contain genes for systems used only in higher animals. This finding is not surprising in cosmic ancestry.
We don't have a clue — Gerard Manning of the Salk Institute, answering the question, "What is a single-celled organism doing with all this communications gear?" We were absolutely stunned. Manning's group is another one studying Monosiga brevicollis, found to contain genes for systems used only in higher animals. This finding is not surprising in cosmic ancestry.
 Gerard Manning et al., "The protist Monosiga brevicollis has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan" [Open Access abstract], doi:10.1073/pnas.0801314105, p 9674-9679 v 105, Proc. Natl. Acad. Sci. USA, 15 Jul 2008. Gerard Manning et al., "The protist Monosiga brevicollis has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan" [Open Access abstract], doi:10.1073/pnas.0801314105, p 9674-9679 v 105, Proc. Natl. Acad. Sci. USA, 15 Jul 2008.
 Can You Hear Me Now?, Salk Institute for Biological Studies, 7 Jul 2008. Can You Hear Me Now?, Salk Institute for Biological Studies, 7 Jul 2008.
 Monosiga brevicollis information from the Joint Genome Institute. Monosiga brevicollis information from the Joint Genome Institute.
 Multicelled animals use a three-part molecular toolkit: a What'sNEW article about the same phenomenon, 4 Jul 2008. Multicelled animals use a three-part molecular toolkit: a What'sNEW article about the same phenomenon, 4 Jul 2008.
 Metazoan Genes Older Than Metazoa? is a related CA webpage. Metazoan Genes Older Than Metazoa? is a related CA webpage.
 8 July 2008 8 July 2008
 A What'sNEW cover page has been added to this website. Now it may take an extra step to get here, but links to NEW articles will not change when they age into the Archives. We hope this will be better for everyone.
A What'sNEW cover page has been added to this website. Now it may take an extra step to get here, but links to NEW articles will not change when they age into the Archives. We hope this will be better for everyone.
 What'sNEW Cover Page — from there, click or wait to be redirected to What'sNEW. What'sNEW Cover Page — from there, click or wait to be redirected to What'sNEW.
 About This Website is a related CA webpage. About This Website is a related CA webpage.
 7 July 2008 7 July 2008
The Design Matrix: A Consilience of Clues, by someone writing as Mike Gene, was recently sent to us. We occasionally read tracts by proponents of Intelligent Design (ID) because our questions about darwinism are often the same as theirs. This book promotes a four-criteria scoring system for deciding whether something was designed "by a mind" or not. Ultimately it gives high probability scores to books, cars, and the genetic code. Surprisingly, the writer never overtly attempts to overturn the scientific approach, and never even mentions the big bang. His brief in support of Michael Behe's irreducible complexity is strictly logical and thought provoking. The text is more informative and less assertive than many others we have seen in support of ID.
The writer argues that ID and darwinian natural selection are not mutually exclusive. Introducing the concept of "Original Mature Design," he explains how the genetic programs for highly-evolved life could be carried forward by more primitive life from the deep past. In support of this concept he cites, as we do, the presence of genes for complex features in primitive organisms without those features. And he mentions several examples of "moonlighting proteins," a phenomenon we knew, but a term we didn't. The hypothesis that very old genetic programs can be activated by darwinian tinkering, also mentioned by Behe, is actually quite close to principles of our own cosmic ancestry. This is interesting. But panspermia is never mentioned.
 We wonder where the writer goes as he probes the past more deeply. Is supernatural intervention required? How does it work? And we are suspicious if someone uses a pseudonym — what is his real ID! Aware of such questions, the writer has a blog wherein he discusses his refusal to answer. "The silence is part of a political strategy itself. This, at least, is how many critics of Intelligent Design think." Right.
We wonder where the writer goes as he probes the past more deeply. Is supernatural intervention required? How does it work? And we are suspicious if someone uses a pseudonym — what is his real ID! Aware of such questions, the writer has a blog wherein he discusses his refusal to answer. "The silence is part of a political strategy itself. This, at least, is how many critics of Intelligent Design think." Right.
We have long complained about the false dichotomy of choices for explaining evolution and the origin-of-life, namely darwinism or creationism/ID. Both camps, comprising 99% of people who discuss the subject, believe that we must explain how life and the genetic programs for higher life forms originated. But if they can originate, there should be credible direct evidence of such processes. Until there is, virtually everyone is trying to explain never-observed phenomena. Without evidence, both camps rely heavily on faith (in the big bang!) to mandate that these origins must occur. And to explain them, both resort to miracles — either incredible unlikelihoods or supernatural intervention.
Cosmic ancestry is a third alternative. It offers a plausible way to explain what we observe, scepticism about what we don't observe, and no need for ongoing miracles. We think it deserves a role in the discussion. Meanwhile, we welcome support for the hypothesis that very old genetic programs are available in Earth's biosphere and can be activated by darwinian tinkering. Panspermia, anyone?
 Mike Gene, The Design Matrix: A Consilience of Clues, ISBN:978-0-9786314-0-6, Arbor Vitae Press, 30 Nov 2007. Mike Gene, The Design Matrix: A Consilience of Clues, ISBN:978-0-9786314-0-6, Arbor Vitae Press, 30 Nov 2007.
 Comparing Darwinism, Creationism/ID and Cosmic Ancestry is a related CA webpage. Comparing Darwinism, Creationism/ID and Cosmic Ancestry is a related CA webpage.
 Evolution vs Creationism is a related CA webpage. Evolution vs Creationism is a related CA webpage.
 Creationism versus Darwinism: A Third Alternative is a related CA online reprint. Creationism versus Darwinism: A Third Alternative is a related CA online reprint.
 Open email to the writer, 7 Jul 2008; and the writer's reply, 20 Jul 2008. Open email to the writer, 7 Jul 2008; and the writer's reply, 20 Jul 2008.
 04 Jul 2008: a three-part molecular toolkit for highly-evolved life carried by more primitive life. 04 Jul 2008: a three-part molecular toolkit for highly-evolved life carried by more primitive life.
 5 July 2008 5 July 2008
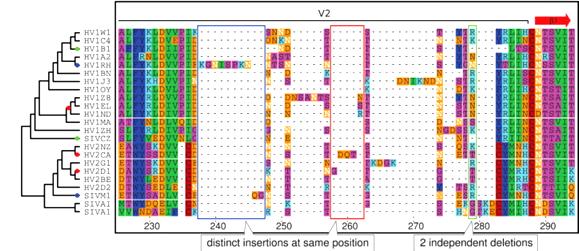 Current methods of gene sequence alignment are error-prone, according to a pair of molecular biologists from the European Bioinformatics Institute in Hinxton, UK. Current methods default to the assumption that similar sequences in the same place are inherited from a common ancestor. This and related assumptions are often wrong and lead to incorrect reconstructions of evolutionary history.
Current methods of gene sequence alignment are error-prone, according to a pair of molecular biologists from the European Bioinformatics Institute in Hinxton, UK. Current methods default to the assumption that similar sequences in the same place are inherited from a common ancestor. This and related assumptions are often wrong and lead to incorrect reconstructions of evolutionary history.
The pair from EBI have devised a "phylogeny-aware" method that is better able to recognize multiple separate insertions at the same location or nearby. For example, in a sequence of amino acid positions in 23 strains of HIV and SIV, older methods find multiple point substitutions and eight independent deletions. In the same sequence, the phylogeny-aware method (illustrated) detects instead seven insertions, two deletions, and fewer point substitutions. Targetted insertions are essential for evolutionary progress in cosmic ancestry. We welcome this clue that they are more common than previously thought.
 Ari Löytynoja and Nick Goldman, "Phylogeny-Aware Gap Placement Prevents Errors in Sequence Alignment and Evolutionary Analysis" [abstract], doi:10.1126/science.1158395, p 1632-1635 v 320, Science, 20 Jun 2008. Ari Löytynoja and Nick Goldman, "Phylogeny-Aware Gap Placement Prevents Errors in Sequence Alignment and Evolutionary Analysis" [abstract], doi:10.1126/science.1158395, p 1632-1635 v 320, Science, 20 Jun 2008.
 Bats and horses are closely related is the conclusion of a study using older methods. Could overlooked insertions point to something more plausible? What'sNEW, 20 Jun 2006. Bats and horses are closely related is the conclusion of a study using older methods. Could overlooked insertions point to something more plausible? What'sNEW, 20 Jun 2006.
 4 July 2008 4 July 2008
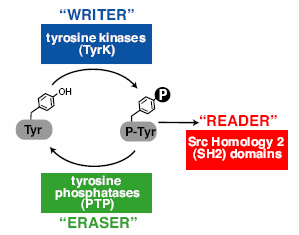 Multicelled animals use a three-part molecular toolkit to mediate phospho-tyrosine signaling. This toolkit enables animals to conduct a number of important communications between their cells, including immune system responses, hormone system stimulation and other crucial functions. "Without these three molecules to help our cells 'write,' 'read' and 'erase' chemical messages between them, our bodies would never be able to conduct the complex tasks needed to survive, such as reproduction, digesting food or even breathing." An international team studied 41 eukaryotic genomes, asking, "How did this system of three components evolve, given their interdependent function?"
Multicelled animals use a three-part molecular toolkit to mediate phospho-tyrosine signaling. This toolkit enables animals to conduct a number of important communications between their cells, including immune system responses, hormone system stimulation and other crucial functions. "Without these three molecules to help our cells 'write,' 'read' and 'erase' chemical messages between them, our bodies would never be able to conduct the complex tasks needed to survive, such as reproduction, digesting food or even breathing." An international team studied 41 eukaryotic genomes, asking, "How did this system of three components evolve, given their interdependent function?" They found that a handful of single-celled eukaryotic species have genes for two of the enzymatic functions, writing and erasing. "This makes sense considering these organisms don't need the tools to communicate between cells since they are made up of only one cell." But why would they need any of them? Well, maybe this incomplete set confers a previously unsuspected benefit, "most likely to deal with limited tyrosine phosphorylation cross-catalyzed by promiscuous Ser/Thr kinases."
 One single-celled species, the choanoflagellate Monosiga brevicollis (illustrated, left), has genes for all three functions. "The researchers conclude that the presence of the full three-component signaling system may have played a role in the development of metazoan organisms.... Probably there was an ancestor to these organisms that first developed these chemicals." But what use are they to one-celled choanoflagellates? "[They] may potentially give cells a wide range of communication possibilities, including uses within single cells. ...There's a certain amount of signaling you can do, and you allocate that apparently for whatever function you want."
One single-celled species, the choanoflagellate Monosiga brevicollis (illustrated, left), has genes for all three functions. "The researchers conclude that the presence of the full three-component signaling system may have played a role in the development of metazoan organisms.... Probably there was an ancestor to these organisms that first developed these chemicals." But what use are they to one-celled choanoflagellates? "[They] may potentially give cells a wide range of communication possibilities, including uses within single cells. ...There's a certain amount of signaling you can do, and you allocate that apparently for whatever function you want."
We commend this analysis of phospho-tyrosine signaling in metazoa. But please notice that it does not even claim to account for the origin of the three-part toolkit. The parts are only observed to exist already in single-celled eukaryotes with no obvious use for them. Actually, finding genes with unexplained origins and no plausible initial purpose is the usual result in studies seeking the origins of genes. Under these circumstances one might expect darwinists to step back and reconsider some of their assumptions. Not yet, apparently.
 David Pincus et al., "Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages" [Open Access abstract], doi:10.1073/pnas.0803161105, p 9680-9684 v 105, Proc. Natl. Acad. Sci. USA, 15 Jul (online 3 Jul) 2008. David Pincus et al., "Evolution of the phospho-tyrosine signaling machinery in premetazoan lineages" [Open Access abstract], doi:10.1073/pnas.0803161105, p 9680-9684 v 105, Proc. Natl. Acad. Sci. USA, 15 Jul (online 3 Jul) 2008.
 Crossed (Evolutionary) Signals?, Press Release 08-113, National Science Foundation, 1 Jul 2008. Crossed (Evolutionary) Signals?, Press Release 08-113, National Science Foundation, 1 Jul 2008.
 Metazoan Genes Older Than Metazoa? is a related CA webpage. Metazoan Genes Older Than Metazoa? is a related CA webpage.
 More metazoan genes came before metazoa – a study of Monosiga brevicollis with a similar finding, What'sNEW, 16 Feb 2008. More metazoan genes came before metazoa – a study of Monosiga brevicollis with a similar finding, What'sNEW, 16 Feb 2008.
 More genes seem to precede the need for themselves – genes for multi-celled functions in choanoflagellates, What'sNEW, 20 Jul 2003. More genes seem to precede the need for themselves – genes for multi-celled functions in choanoflagellates, What'sNEW, 20 Jul 2003.
 A gene needed for multcellularity... – a receptor tyrosine kinase in choanoflagellates, What'sNEW, 21 Dec 2001. A gene needed for multcellularity... – a receptor tyrosine kinase in choanoflagellates, What'sNEW, 21 Dec 2001.
 Thanks, Stan Franklin. Thanks, Stan Franklin.
 25 June 2008 25 June 2008
Vertebrate and jellyfish eyes use similar genes. Both lineages probably recruited them independently. An international research team writes: Combined, the available data favor the possibility that vertebrate and cubozoan eyes arose by independent recruitment of orthologous genes during evolution. Otherwise, if they were inherited from a common ancestor (the default assumption), many other lineages apparently lost them. This loss would violate the principle that evolution preserves advantageous features.
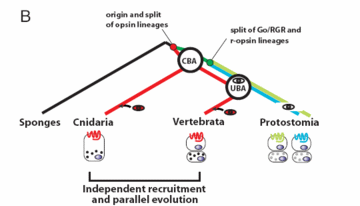 Actually, neither scenario supports the darwinian paradigm of gradual program composition. If the genes were independently recruited, their ultimate source is not accounted for in either vertebrate or jellyfish lineages. But if they come from common ancestry, the genes appear to wholly precede the first appearance of the features they encode.
Actually, neither scenario supports the darwinian paradigm of gradual program composition. If the genes were independently recruited, their ultimate source is not accounted for in either vertebrate or jellyfish lineages. But if they come from common ancestry, the genes appear to wholly precede the first appearance of the features they encode.
For other possibilities, could convergent darwinian evolution produce the same genes multiple times? The similarities of the subject genes, apparently, make this implausible. Or, could the genes have evolved in the vertebrate lineage according to darwin, and then been acquired by jellyfish? After all, we are looking at today's jellyfish. The genomic-historical-reconstruction method suffers from this ambiguity.
The research team further notes that recruitment of genes by widely diverse species is not uncommon: …Entire regulatory circuits can be co-opted for development of different cell types, tissues, or organs. For example, the Pax–Six–Eya–Dach gene regulatory network has a fundamental role in Drosophila visual system development but is also used for specification of muscle cells or placodes in vertebrates. This is how evolution is expected to work in cosmic ancestry — genetic programs are not gradually composed, but acquired. If regulatory genes are included in the acquisitions, we are not surprised.
 Zbynek Kozmik et al., "Assembly of the cnidarian camera-type eye from vertebrate-like components" [abstract], doi:10.1073/pnas.0800388105, p 8989-8993 v 105, Proc. Natl. Acad. Sci. USA, 1 Jul (online 24 Jun) 2008. Zbynek Kozmik et al., "Assembly of the cnidarian camera-type eye from vertebrate-like components" [abstract], doi:10.1073/pnas.0800388105, p 8989-8993 v 105, Proc. Natl. Acad. Sci. USA, 1 Jul (online 24 Jun) 2008.
 The key to early eye evolution? is an earlier What'sNEW article about jellyfish eyes, 21 May 2005. The key to early eye evolution? is an earlier What'sNEW article about jellyfish eyes, 21 May 2005.
 Neo-Darwinism... is a related CA webpage with a discussion of eyes under Coordinating Genes. Neo-Darwinism... is a related CA webpage with a discussion of eyes under Coordinating Genes.
 Viruses and Other Gene Transfer Mechanisms is our main CA webpage about gene acquisition Viruses and Other Gene Transfer Mechanisms is our main CA webpage about gene acquisition
[ What'sNEW about HGT What'sNEW about HGT  ] ]
 Testing Darwinism versus Cosmic Ancestry is a CA webpage comparing methods for testing evolution. Testing Darwinism versus Cosmic Ancestry is a CA webpage comparing methods for testing evolution.

 24 June 2008 24 June 2008
...We have found proof that this ...is really water ice — Peter Smith, Principal Investigator for the Phoenix mission to Mars. The key new evidence is that chunks of bright material exposed by digging on June 15 and still present on June 16 had vaporized by June 19. The bright chunks are visible in the bottom left of the sol 20 photo, in the deepest, shadowed part of the imprint of the scoop. They are missing on sol 24.
 NASA Phoenix Mars Lander Confirms Frozen Water, NASA, 20 Jun 2008. NASA Phoenix Mars Lander Confirms Frozen Water, NASA, 20 Jun 2008.
 Life on Mars! is a related CA webpage. Life on Mars! is a related CA webpage.
 Thanks, Richard Hoover. Thanks, Richard Hoover.
 21 June 2008 21 June 2008
Panspermia is considered in depth in a new web article that gives a brief history of the idea, mentions various versions, and discusses the evidence in a well-balanced manner. Among other matters of interest is word that Arizona State University astrobiologist Paul Davies is concerned about the software aspect of the origin-of-life problem. In response to renewed claims that organics from space could kickstart life on Earth, he says, "Life is all about information...." Good.

 Panspermia Theory for Life's Origins Gets Boost..., MensNewsDaily.com (+Newswax), 20 Jun 2008. Panspermia Theory for Life's Origins Gets Boost..., MensNewsDaily.com (+Newswax), 20 Jun 2008.
 Introduction: More Than Panspermia is a related CA webpage. Introduction: More Than Panspermia is a related CA webpage.
 The RNA World is our main webpage about origin-of-life theories. The RNA World is our main webpage about origin-of-life theories.
 Organic compounds were present... is our What'sNEW story about the recent discovery that renewed the "kickstarting" discussion, 14 Jun 2008. Organic compounds were present... is our What'sNEW story about the recent discovery that renewed the "kickstarting" discussion, 14 Jun 2008.
 Thanks, Google Alerts. Thanks, Google Alerts.
 21 June 2008 21 June 2008
Another attempt to advance the Evolution Prize is posted as a challenge on Innocentive.com — "Is Open-Ended Evolutionary Innovation in a Quarantined System Possible?" The Astrobiology Research Trust sponsors the $20,000 award. A discussion forum for the challenge is on Google Groups.

 Innocentive.com — register to view challenge 6409250, offered thru 13 Sep 2008. Innocentive.com — register to view challenge 6409250, offered thru 13 Sep 2008.
 Open-Ended Evolutionary Innovation / Quarantined Syst. — the discussion forum at Google Groups. Open-Ended Evolutionary Innovation / Quarantined Syst. — the discussion forum at Google Groups.
 The Astrobiology Research Trust, established 22 Apr 2002. The Astrobiology Research Trust, established 22 Apr 2002.
 The Evolution Prize, website established Jun 2006. The Evolution Prize, website established Jun 2006.
 Cornelia Dean, "If You Have a Problem, Ask Everyone" [text], The New York Times, 22 Jul 2008. Cornelia Dean, "If You Have a Problem, Ask Everyone" [text], The New York Times, 22 Jul 2008.
 The Evolution Prize... is a related CA webpage, posted 15 Apr 2006. The Evolution Prize... is a related CA webpage, posted 15 Apr 2006.
 14 June 2008 14 June 2008
Organic compounds were present in the early solar system, according to an international collaboration studying the Murchison meteorite. Nine astrobiologists from the The Netherlands, UK and USA used isotopic analysis to determine that uracil and xanthine in the meteorite are not Earthly contaminants. Uracil is a component of RNA, and xanthine (shown in diagram) is an intermediate in the synthesis of other biological molecules. The study suggests that these and other similar products, delivered by meteorites, "would have enriched the prebiotic organic inventory necessary for life to assemble on the early Earth." Could they even be the remnants of prior life?
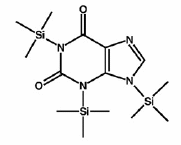
 Zita Martins et al., "Extraterrestrial nucleobases in the Murchison meteorite" [pdf | arXiv.org abstract], doi:10.1016/j.epsl.2008.03.026, p 130-136 v 270, Earth and Planetary Science Letters, 15 Jun (online 20 Mar) 2008. Zita Martins et al., "Extraterrestrial nucleobases in the Murchison meteorite" [pdf | arXiv.org abstract], doi:10.1016/j.epsl.2008.03.026, p 130-136 v 270, Earth and Planetary Science Letters, 15 Jun (online 20 Mar) 2008.
 Were Meteorites the Origin of Life on Earth?, by JR Minkel, Scientific American, 16 Jun 2008. Were Meteorites the Origin of Life on Earth?, by JR Minkel, Scientific American, 16 Jun 2008.
 Life Cooked Up in Outer Space?, by Phil Berardelli, ScienceNOW Daily News, 16 Jun 2008. Life Cooked Up in Outer Space?, by Phil Berardelli, ScienceNOW Daily News, 16 Jun 2008.
 ...Parts of earliest genetic material may have come from the stars, PhysOrg.com, 13 Jun 2008. ...Parts of earliest genetic material may have come from the stars, PhysOrg.com, 13 Jun 2008.
 ...Life's Raw Material Came from Space, by Clara Moskowitz, Space.com, 13 Jun 2008. ...Life's Raw Material Came from Space, by Clara Moskowitz, Space.com, 13 Jun 2008.
 Amino Acid Asymmetry in the Murchison Meteorite! is a related CA webpage, posted 14 Feb 1997. Amino Acid Asymmetry in the Murchison Meteorite! is a related CA webpage, posted 14 Feb 1997.
 Fossilized Life Forms in the Murchison Meteorite is a related CA webpage, posted 29 Jul 1997. Fossilized Life Forms in the Murchison Meteorite is a related CA webpage, posted 29 Jul 1997.
 Fossilized Bacteria in Murchison and Efremovka is a related CA webpage, posted 27 Jan 2000. Fossilized Bacteria in Murchison and Efremovka is a related CA webpage, posted 27 Jan 2000.
 Thanks, Zita Martins, Stan Franklin, Jacob Navia, Jim Galasyn, Kevin Hatfield, Larry Klaes and Google Alerts. And Charlie Newman? Thanks, Zita Martins, Stan Franklin, Jacob Navia, Jim Galasyn, Kevin Hatfield, Larry Klaes and Google Alerts. And Charlie Newman?
 11 June 2008 11 June 2008
 We will see organics, for sure, because we're bringing them, says Aaron Zent of NASA's Ames Research Center, talking about the organics detector that the Phoenix lander just delivered to the surface of Mars. TEGA, the Thermal and Evolved Gas Analyzer, can detect much lower concentrations of organics than the instruments sent to Mars with the Viking missions that landed in 1976 and 1977, which detected no organics. However, apparently, TEGA is so sensitive that it may end up sensing only contamination from Earth.
We will see organics, for sure, because we're bringing them, says Aaron Zent of NASA's Ames Research Center, talking about the organics detector that the Phoenix lander just delivered to the surface of Mars. TEGA, the Thermal and Evolved Gas Analyzer, can detect much lower concentrations of organics than the instruments sent to Mars with the Viking missions that landed in 1976 and 1977, which detected no organics. However, apparently, TEGA is so sensitive that it may end up sensing only contamination from Earth. Of course the Phoenix team would dispute the suggestion that TEGA is unreliable. But in general, biological contamination is pervasive and extremely difficult to prevent. Only lately has the true magnitude of this problem been recognized. Even when non-contamination is the first priority in controlled laboratory experiments, contamination still happens. To achieve reliable results in biology, foolproof protocols are needed. Can such protocols be designed? In a related issue, for testing the range of evolution in closed systems, would nonbiological media such as computer models be preferable?
 Eric Hand, "'Dandruff' could contaminate Phoenix landing site" [html], doi:10.1038/news.2008.878, Nature, online 6 Jun 2008. Eric Hand, "'Dandruff' could contaminate Phoenix landing site" [html], doi:10.1038/news.2008.878, Nature, online 6 Jun 2008.
 Life on Mars! is a related CA webpage with more about the Viking missions. Life on Mars! is a related CA webpage with more about the Viking missions.
 Testing Darwinism versus Cosmic Ancestry is a CA webpage comparing methods for testing evolution. Testing Darwinism versus Cosmic Ancestry is a CA webpage comparing methods for testing evolution.
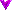 Cloned bacteria... is the example that we have in mind illustrating laboratory contamination, 5 Jun 2008. Cloned bacteria... is the example that we have in mind illustrating laboratory contamination, 5 Jun 2008.
 5 June 2008 5 June 2008
Cloned bacteria evolved an unexpected feature in a long-running experiment led by Richard Lenski at Michigan State University. After some 31,500 generations, a population of E. coli suddenly began to metabolize citrate aerobically. The inability to do so is a defining characteristic of this species, and this rare adaptation looks like a significant innovation. The Long-Term Evolution Experiment (LTEE), started in 1988, has now run for more than 44,000 generations. The observed mutation rate is apparently sufficient to have supplied for testing, many times, every typical one-step mutation available to the 4.6 million base-pair genome. And frozen samples that remain viable are taken every 500 generations, so it is possible to reanalyze prior stages of the process. The system is uniquely useful for studying evolution.
 The research team was especially interested to know whether the latest adaptation "required an unusually rare mutation or, alternatively, was historically contingent and depended on the prior evolution of a certain genetic background." Using the frozen samples, they decided that historical contingency was the answer.
The research team was especially interested to know whether the latest adaptation "required an unusually rare mutation or, alternatively, was historically contingent and depended on the prior evolution of a certain genetic background." Using the frozen samples, they decided that historical contingency was the answer.
They then ask, "What is the genetic basis of this evolutionary innovation?" And they acknowledge, "We anticipate that identifying the potentiating mutation will be especially challenging, however, because its only known phenotype is to increase the rate of production of certain mutants that are themselves extremely rare." While there are difficulties, we feel that closed-system research like this is exactly what is needed for truly understanding how evolution works. We are pleased that MSU is doing it.
Of course, understanding how evolution works is a primary concern of our own as well. Specifically, we want to know if mutations in closed systems like this one can write new genetic programs or not. If so, as mainstream science assumes, the darwinian paradigm would be sufficient to explain the evolution of life on Earth, with no need for input from elsewhere. If not, as we suspect, darwinian evolution would be limited to exploring the capabilities of the programs that are provided, initially or eventually. We believe ours is a more conservative position, and that the mainstream proposition amounts to an extraordinary claim. Therefore we view the evidence with unusual scrutiny. The MSU researchers acknowledge the possibility that "some complex mutation, or multiple mutations, activated cryptic genes that jointly expressed a citrate transporter." They also mention that contamination occasionally happens in the experiment, although the strain which underwent the studied adaptation is not a contaminant. However, E. coli "from agricultural and clinical settings ...harbor plasmids, presumably acquired from other species, that encode citrate transporters." Could a non-surviving contaminant have introduced similar plasmids in this case? These possibilities — cryptic genes and contamination — undermine the argument that mutations wrote a new program this time. Perhaps more experiments and analysis will uphold the darwinian position. If so, we think Richard Lenski's approach will be the one that succeeds. Meanwhile, the capability of darwinian evolution to write new programs in a closed system has not been clearly demonstrated.
 Zachary D. Blount, Christina Z. Borland, and Richard E. Lenski, "Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli" [abstract], doi:10.1073/pnas.0803151105, p 7899-7906 v 105, Proc. Nat. Acad. Sci. USA, 10 Jun 2008. Zachary D. Blount, Christina Z. Borland, and Richard E. Lenski, "Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli" [abstract], doi:10.1073/pnas.0803151105, p 7899-7906 v 105, Proc. Nat. Acad. Sci. USA, 10 Jun 2008.
 Bacteria make major evolutionary shift in the lab by Bob Holmes, NewScientist, 9 Jun 2008. Bacteria make major evolutionary shift in the lab by Bob Holmes, NewScientist, 9 Jun 2008.
 ...Is Evolutionary Progress in a Closed System Possible is a CA webpage with discussion and links about Lenski's experiments. ...Is Evolutionary Progress in a Closed System Possible is a CA webpage with discussion and links about Lenski's experiments.
 Testing Darwinism versus Cosmic Ancestry is a CA webpage explaining why we like Lenski's method. Testing Darwinism versus Cosmic Ancestry is a CA webpage explaining why we like Lenski's method.
  20 Sep 2012: Richard Lenski's research group has analysed the evolution of aerobic citrate metabolism.... 20 Sep 2012: Richard Lenski's research group has analysed the evolution of aerobic citrate metabolism....
  4 June 2008 4 June 2008
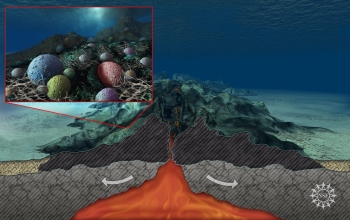
Prokaryotic life in Earth's deep dark ocean, subsisting on rock, has been observed to be far more abundant and diverse than expected. "We now know that this remote region is teeming with microbes, more so than anyone had guessed," said David Garrison, director of the National Science Foundation's Biological Oceanography Program. Would similar life be able to to subsist on the bottom of deep dark oceans on other moons or planets such as, possibly, Ganymende, Enceladus, Europa, or Callisto? Why not?
 Bacteria 'Feed' on Earth's Ocean-Bottom Crust, Press Release 08-086, National Science Foundation, 28 May 2008. Bacteria 'Feed' on Earth's Ocean-Bottom Crust, Press Release 08-086, National Science Foundation, 28 May 2008.
 Rock eating deep sea baterica! [sic], nature.com, 29 May 2008. Rock eating deep sea baterica! [sic], nature.com, 29 May 2008.
 Bacteria... is a related CA webpage. Bacteria... is a related CA webpage.
 Life on Europa or Other Moons? is a CA webpage with related links. Life on Europa or Other Moons? is a CA webpage with related links.
 Erwan G. Roussel et al., "Extending the Sub-Sea-Floor Biosphere" [abstract], doi:10.1126/science.1154545, p 1046 v 320, Science, 23 May 2008. "...Evidence for low concentrations of living prokaryotic cells in the deepest (1626 meters below the sea floor), oldest (111 million years old), and potentially hottest (~100°C) marine sediments investigated." Erwan G. Roussel et al., "Extending the Sub-Sea-Floor Biosphere" [abstract], doi:10.1126/science.1154545, p 1046 v 320, Science, 23 May 2008. "...Evidence for low concentrations of living prokaryotic cells in the deepest (1626 meters below the sea floor), oldest (111 million years old), and potentially hottest (~100°C) marine sediments investigated."
 3 June 2008 3 June 2008
After decades of effort, some scientists begin to despair of explaining the universe.
 Dennis Overbye, "Dark, Perhaps Forever" [html], p D1,D4, The New York Times, 3 Jun 2008. Dennis Overbye, "Dark, Perhaps Forever" [html], p D1,D4, The New York Times, 3 Jun 2008.
 The End and the Big Bang is a related CA webpage. The End and the Big Bang is a related CA webpage.
 Thanks, Robert Pugh. Thanks, Robert Pugh.
|
 We don't have a clue — Gerard Manning of the Salk Institute, answering the question, "What is a single-celled organism doing with all this communications gear?" We were absolutely stunned. Manning's group is another one studying Monosiga brevicollis, found to contain genes for systems used only in higher animals. This finding is not surprising in cosmic ancestry.
We don't have a clue — Gerard Manning of the Salk Institute, answering the question, "What is a single-celled organism doing with all this communications gear?" We were absolutely stunned. Manning's group is another one studying Monosiga brevicollis, found to contain genes for systems used only in higher animals. This finding is not surprising in cosmic ancestry. A What'sNEW cover page has been added to this website. Now it may take an extra step to get here, but links to NEW articles will not change when they age into the Archives. We hope this will be better for everyone.
A What'sNEW cover page has been added to this website. Now it may take an extra step to get here, but links to NEW articles will not change when they age into the Archives. We hope this will be better for everyone.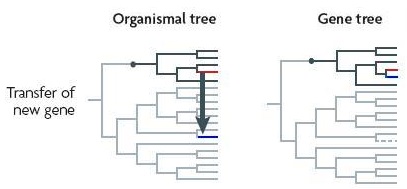
 We wonder where the writer goes as he probes the past more deeply. Is supernatural intervention required? How does it work? And we are suspicious if someone uses a pseudonym — what is his real ID! Aware of such questions, the writer has a blog wherein he discusses his refusal to answer. "The silence is part of a political strategy itself. This, at least, is how many critics of Intelligent Design think." Right.
We wonder where the writer goes as he probes the past more deeply. Is supernatural intervention required? How does it work? And we are suspicious if someone uses a pseudonym — what is his real ID! Aware of such questions, the writer has a blog wherein he discusses his refusal to answer. "The silence is part of a political strategy itself. This, at least, is how many critics of Intelligent Design think." Right. Current methods of gene sequence alignment are error-prone, according to a pair of molecular biologists from the European Bioinformatics Institute in Hinxton, UK. Current methods default to the assumption that similar sequences in the same place are inherited from a common ancestor. This and related assumptions are often wrong and lead to incorrect reconstructions of evolutionary history.
Current methods of gene sequence alignment are error-prone, according to a pair of molecular biologists from the European Bioinformatics Institute in Hinxton, UK. Current methods default to the assumption that similar sequences in the same place are inherited from a common ancestor. This and related assumptions are often wrong and lead to incorrect reconstructions of evolutionary history.
 Multicelled animals use a three-part molecular toolkit to mediate phospho-tyrosine signaling. This toolkit enables animals to conduct a number of important communications between their cells, including immune system responses, hormone system stimulation and other crucial functions. "Without these three molecules to help our cells 'write,' 'read' and 'erase' chemical messages between them, our bodies would never be able to conduct the complex tasks needed to survive, such as reproduction, digesting food or even breathing." An international team studied 41 eukaryotic genomes, asking, "How did this system of three components evolve, given their interdependent function?"
Multicelled animals use a three-part molecular toolkit to mediate phospho-tyrosine signaling. This toolkit enables animals to conduct a number of important communications between their cells, including immune system responses, hormone system stimulation and other crucial functions. "Without these three molecules to help our cells 'write,' 'read' and 'erase' chemical messages between them, our bodies would never be able to conduct the complex tasks needed to survive, such as reproduction, digesting food or even breathing." An international team studied 41 eukaryotic genomes, asking, "How did this system of three components evolve, given their interdependent function?" One single-celled species, the choanoflagellate Monosiga brevicollis (illustrated, left), has genes for all three functions. "The researchers conclude that the presence of the full three-component signaling system may have played a role in the development of metazoan organisms.... Probably there was an ancestor to these organisms that first developed these chemicals." But what use are they to one-celled choanoflagellates? "[They] may potentially give cells a wide range of communication possibilities, including uses within single cells. ...There's a certain amount of signaling you can do, and you allocate that apparently for whatever function you want."
One single-celled species, the choanoflagellate Monosiga brevicollis (illustrated, left), has genes for all three functions. "The researchers conclude that the presence of the full three-component signaling system may have played a role in the development of metazoan organisms.... Probably there was an ancestor to these organisms that first developed these chemicals." But what use are they to one-celled choanoflagellates? "[They] may potentially give cells a wide range of communication possibilities, including uses within single cells. ...There's a certain amount of signaling you can do, and you allocate that apparently for whatever function you want." Actually, neither scenario supports the darwinian paradigm of gradual program composition. If the genes were independently recruited, their ultimate source is not accounted for in either vertebrate or jellyfish lineages. But if they come from common ancestry, the genes appear to wholly precede the first appearance of the features they encode.
Actually, neither scenario supports the darwinian paradigm of gradual program composition. If the genes were independently recruited, their ultimate source is not accounted for in either vertebrate or jellyfish lineages. But if they come from common ancestry, the genes appear to wholly precede the first appearance of the features they encode.


 We will see organics, for sure, because we're bringing them, says Aaron Zent of NASA's Ames Research Center, talking about the organics detector that the Phoenix lander just delivered to the surface of Mars. TEGA, the Thermal and Evolved Gas Analyzer, can detect much lower concentrations of organics than the instruments sent to Mars with the Viking missions that landed in 1976 and 1977, which detected no organics. However, apparently, TEGA is so sensitive that it may end up sensing only contamination from Earth.
We will see organics, for sure, because we're bringing them, says Aaron Zent of NASA's Ames Research Center, talking about the organics detector that the Phoenix lander just delivered to the surface of Mars. TEGA, the Thermal and Evolved Gas Analyzer, can detect much lower concentrations of organics than the instruments sent to Mars with the Viking missions that landed in 1976 and 1977, which detected no organics. However, apparently, TEGA is so sensitive that it may end up sensing only contamination from Earth. The research team was especially interested to know whether the latest adaptation "required an unusually rare mutation or, alternatively, was historically contingent and depended on the prior evolution of a certain genetic background." Using the frozen samples, they decided that historical contingency was the answer.
The research team was especially interested to know whether the latest adaptation "required an unusually rare mutation or, alternatively, was historically contingent and depended on the prior evolution of a certain genetic background." Using the frozen samples, they decided that historical contingency was the answer.